On Thursday April 17, Bin Wu, associate professor in the department of Biophysics and Biophysical Chemistry at the School of Medicine, visited Homewood Campus to deliver a talk titled “Imaging translation of single mRNAs in live cells” as part of the Department of Biology’s Seminar Series. Wu discussed his lab’s recent work using single-molecule imaging techniques to visualize the mechanisms of messenger RNA (mRNA) translation in real time.
Translation is a fundamental biological process of converting mRNA information into proteins. As part of the central dogma of molecular biology, translation particularly interests Wu because it plays a key role in determining how genetic information is expressed, yet many of the mechanisms involved in the process remain poorly understood.
“We’re interested in how cells regulate expression in space and time,” Wu stated, emphasizing the importance of fine-tuning protein production. “To do that, we’ve [developed] single molecule approaches to visualize the transcription, transport, translation and degradation of genetic information in cells.”
Wu’s lab has pioneered many innovative, state-of-the-art methods to study single molecule nucleic acid biology, including very fast Clustered Regularly Interspaced Short Palindromic Repeats (vfCRISPR) and Rapid Inducible Decay of RNA (RIDR). For this talk, Wu focused on how his lab is studying translational dynamics in single mRNAs over extended periods of time using an imaging system called single-molecule imaging of nascent peptides (SINAPS), which he co-developed as a postdoc student.
One round of translation into protein can take several minutes, which — prior to the development of SINAPS — made it challenging to continuously image a single mRNA molecule that is freely diffusing in the cell for the entire time frame.
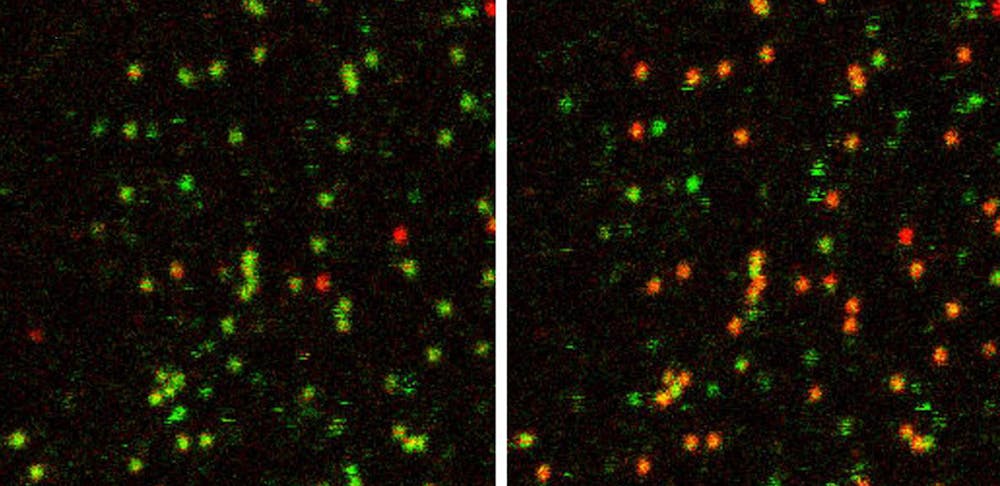
“In order to track the translation of one RNA for an extended period of time, you would have had to track an mRNA for many hours... we adapted [SINAPS] to tether the mRNA to the bottom of the plasma membrane,” Wu described.
Using a system of modified hairpin loops in the untranslated region (3’-UTR) of the mRNA that were either fluorescently tagged or anchored to membrane proteins, Wu’s team was able to track mRNAs for hours, enabling them to observe individual mRNAs undergoing multiple rounds of translation.
“As a result, we were able to see that the translation of mRNA turns on and off — it does not show a constant translation signal,” Wu noted.
Instead, translation occurs in “bursts,” similar to the movement of incoming and receding waves. Further experiments elucidated the mechanisms responsible for switching mRNAs between active and inactive states for translation. Wu’s lab identified factors in the 5’-UTR that regulate both the timing and amount of translation initiation, including hairpin sizes and translation factor sequences that alter the mRNA’s secondary structure and the ability of ribosomes to locate the start codon.
Wu characterized the “bursty” nature of translation in terms of both frequency (the timing of initiation) and amplitude (the amount of newly formed peptides from ribosome performing translation). Similar bursting behavior is also observed in transcription of DNA into RNA, with well-characterized transcription factor and enhancer-promoter interactions that may be analogous to translational regulation.
Wu then shifted to a discussion of his lab’s investigation into the mechanisms of mRNA quality control using single-molecule imaging.
“For a cell to maintain its proteins properly, you need to be able to test whether the message transcribed is good or bad,” he stated. “However, due to transcription errors, mutations or DNA damage, cells often have [error mRNA] messages that need to be detected.”
Wu went on to explain that problematic mRNA sequences can cause ribosomes to stall, which results in collisions when ribosomes translating upstream of the same mRNA reach the stalled ribosome. Detecting and resolving collided ribosomes is an important quality-control response, involving ZNF598 — an E3 ubiquitin ligase that labels the ribosome complex for degradation.
Wu’s team used long stalling poly-A sequences to create a blockade of ribosomes at the 3’ end of mRNAs in order to analyze the time required for ZNF598 to clear the blockade, enabling them study ribosome collision-triggered mRNA quality control in vivo.
Indeed, when ZNF598 was overexpressed, collisions were resolved faster, and conversely, knocking down ZNF598 expression resulted in much slower clearance. Interestingly, Wu’s team observed that several ZNFs could bind to a train of collisions, but they never saturated all collision sites. To Wu, these results suggested that ZNF598 — due to its low abundance — was the rate-limiting factor for triggering ribosomal quality control. Furthermore, he hypothesized that this mechanism limits ZNF intervention only for extended periods of collision; transient collisions are resolved without needing ZNF.
By characterizing how ZNF598 responds to ribosome collisions, Wu’s findings help facilitate an improved understanding of mRNA quality control via ribosome dynamics. Additionally, the group’s development of tools to visualize single particles during translation can be applied to other dynamic cellular processes like DNA replication, gene transcription and more, allowing scientists to answer fundamental remaining questions about basic cellular processes.