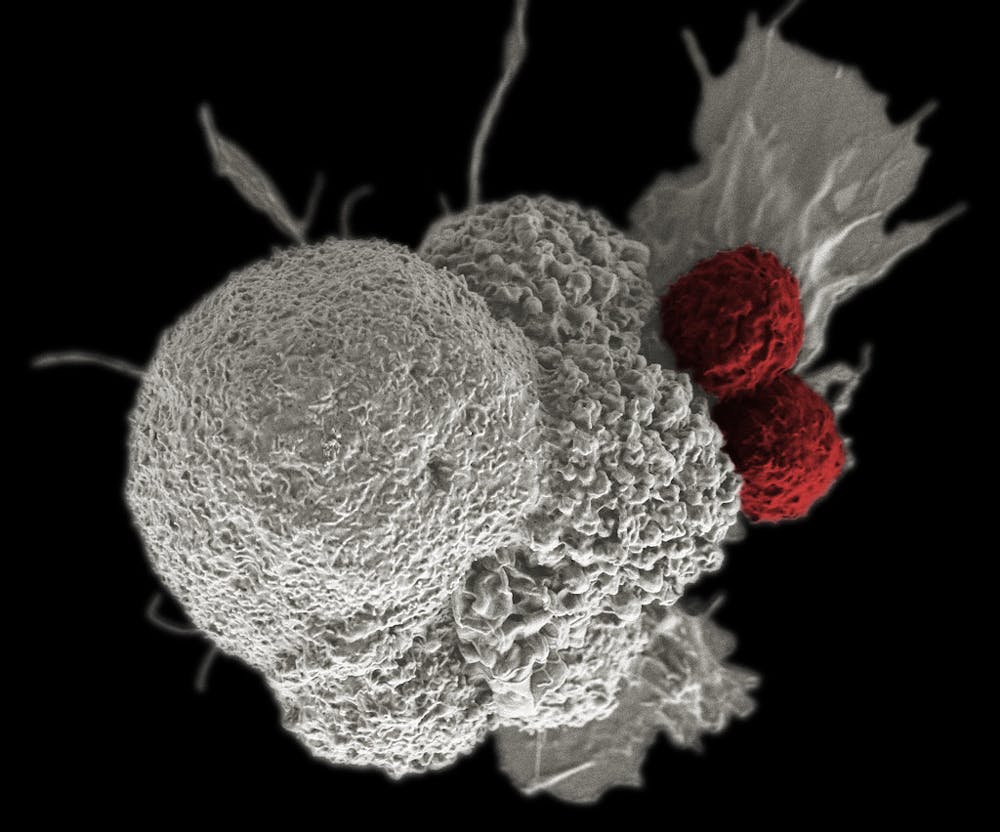
Immunotherapy holds great promise for the future of cancer treatment. By harnessing a patient’s own immune system to target cancerous cells, cancer treatment can be tailored to an individual’s specific cancer type — allowing for more personalized treatment. One key avenue of current research involves studying cellular organization within tumors to understand the role of tumor-associated macrophages: white blood cells closely associated with the tumor microenvironment (TME) and are thought to support tumor growth and invasiveness.
Researchers from the Sidney Kimmel Comprehensive Cancer Center conducted an in-depth spatial analysis of the breast cancer TME to investigate the relationships between immune cells and breast cancer cells that are responsible for initiating metastasis — the progressive spreading of cancerous growths to areas physically distant from a primary site. This research represents interdisciplinary work involving multiple professors at Hopkins.
In particular, this study involved collaboration with and drew heavily from work previously done by primary author Dr. Eloïse M. Grasset, who now works at the Center for Research in Cancerology and Immunology Nantes-Angers in France, and Dr. Andrew Ewald — a professor of Biomedical Engineering and Oncology and co-leader of the Cancer Invasion and Metastasis Program at the Kimmel Cancer Center. Their most recent study used imaging mass cytometry (IMC) to study cell-cell interactions in the TME. IMC is a combination of two traditionally used techniques: elemental mass spectrometry and flow cytometry.
Traditional flow cytometry uses fluorescent probes attached to antibodies to label cells; however, the use of fluorescence can often lead to spillover effects from different fluorophores, making it more difficult to identify and analyze specific cell subtypes. In contrast, mass cytometry uses antibodies that bind to rare earth metal “reporters.” These reporters are isotopically enriched, meaning that 99.9% of isotopes that exist of the reporter exist at the same molecular weight. Using a modified mass spectrometry machine, researchers can detect these very specific masses at single-cell resolution. The quantity of reporter ions then acts as a way to measure molecular expression and limit signal overlap between the different parameters.
“Even very experienced flow cytometrists might get you about 20 parameters...whereas [with] mass cytometry, it’s pretty easy to put together a panel of 30 or 40 parameters. Now we use the same technology on tissues, and that’s the imaging part of it,” explained Dr. Won Jin Ho — a senior author on the study and an assistant professor of oncology — in an interview with The News-Letter. Ho is a physician-scientist at the Kimmel Cancer Center and the director of the center’s mass cytometry facility.
After performing IMC on biospecimens of 24 breast cancer patients, the data was then analyzed using existing bioinformatics pipelines. The researchers were able to cluster and annotate specific subtypes of cells based on the biomarkers they displayed. This single-cell resolution helped them understand how cells were spatially coordinated — in other words, what immune cell subtypes were touching cells responsible for cancer progression?
Data analysis revealed that a small subset (about 5%) of macrophages were consistently found in close proximity to metastasis-promoting breast cancer cells. These macrophages also expressed a distinct set of biomarkers (specific molecules that are indicative of a particular cell type), allowing the researchers to categorize them as a distinct subtype of macrophages compared to other immune cells found in the TME. The data-driven approach previously described eliminated bias during cell clustering, ensuring that the conclusions of the study were representative of the data collected and not affected by the analytical methods used to process the data.
“There’s no bias in trying to come up with different groups of cells; you just input the data and see how the data drives the similarities across different cell types,” he said. “And then we can annotate based on the expression profiles of what these cells actually represent.”
Image segmentation analysis — where an image is partitioned into groups of pixels to easily process different quantitative markers — can often lead to the presence of artifacts which can affect the quality of analysis. To ensure these artifacts didn’t impact their analysis, researchers for this study further cross-validated their conclusions by analyzing a publicly available, independent dataset that dissociated the tumor into individual cells, creating a true single-cell dataset.
“By doing this cross-validation which is very important in bioinformatics, we were able to have extra confidence that the data really identified specific phenotypes that would be interesting,” Ho concluded.
Ho also provided insights on what the interactions between immune cells and breast cells could potentially be and how they contributed to cancer progression. One hypothesis he provided was cytokine-mediated interactions. Oftentimes, cancer cells secrete immunosuppressive cytokines — specialized signaling proteins involved in immune system function — that can impact nearby macrophages in a paracrine manner.
Another hypothesis Ho suggested relates to the high expression of arginase-1 (ARG1) in breast cancer cells. ARG1 has been historically proven to play a role in tumorigenesis and metastasis, and some of the cells studied near metastasis-initiating breast cancer cells in this study had elevated ARG1 expression.
“Some of these macrophages have high arginase-1 expression, and this represents some change in how the environment will look to the cancer in terms of its metabolic activity. Is there anything related to the metabolic state of these cancer cells when they are close to these macrophages?” he hypothesized.
Moving forward, the researchers have several avenues through which they hope to study these macrophage-tumor cell interactions. One of the most promising involves organoid research, where tumor cells are harvested and cultured in 3D environments known as organoids. By co-culturing these organoids with traditional elements of the breast cancer TME, researchers can look at specific cell-cell interactions to see how the subtype of macrophages they identified contributes to metastasis initiation by breast cancer cells.
“When you use cancer cell organoids, you can emulate or capture some of their behavior that resembles what’s happening in vivo... you can understand how invasive they might look and how they behave... and it might actually resemble what’s happening inside the body,” Ho explained.
Ho also provided perspective on how this study could have implications for the bigger-picture future of immunotherapy. By understanding the interaction between macrophages and breast cancer cells, it could be possible to reprogram immune cells in the TME to curb metastasis.
“It’s a huge actively ongoing effort in the field... metastasis is really the turning point in a patient’s prognosis, so if we can curb metastasis, hopefully we can really improve patient survival,” he concluded.