Jean Fan is an assistant professor leading the JEFworks Lab at The Center for Computational Biology (CCB). In a recent interview with The News-Letter, Fan shared her work and the recent progress of her team in the field of spatial transcriptomics.
Traditional bulk sequencing methods, such as Sanger Sequencing and Next-Generation Sequencing, are well-established technologies for obtaining reads of RNA sequences. However, since these methods treat samples as a bulk mixture of cells, it is difficult to distinguish different cell types within a complex tissue, thus making studying cell organization challenging.
In the recent decade, spatially resolved transcriptomics (SRT) addressed this issue and allowed the exploration of cellular identity and heterogeneity at a single-cell level in tissues. Fan's lab focuses on devising efficient algorithms to handle the immense computational demands that SRT requires given the vast number of genes and cells that can be profiled.
Fan explained the primary application of SRT in understanding the spatial context within cellular systems.
"For example, if we identify a pattern that certain kinds of tumor cells are always found near T cells, this might have a certain implication regarding tumor microenvironment interactions,” she said. “Whereas in another context, we might have a tumor where there aren't any T cells infiltrating but they're all kept on the periphery. In these cases, we first want to be able to determine what these cells are doing, and SRT helps with this by letting us measure what genes they are expressing.”
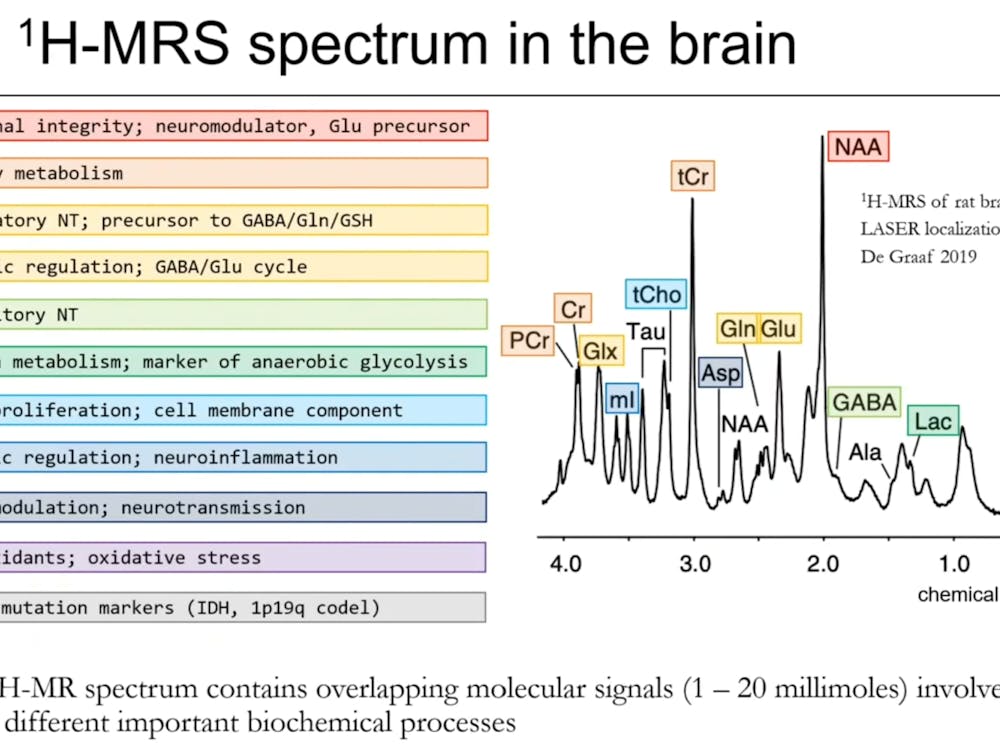
She likened the mechanism of SRT to taking"images" that reveal the diversity of gene expression within a tissue sample.
"We design various probes targeting different genes, which enables us to take pictures of the tissues where different types of genes are highlighted using different combinations of colors," Fan explained. "From these, we could know not only what is expressed but also where exactly within the tissues, and the use of spatial information could help distinguish cellular state and identity."
A recent achievement of Fan's lab is the development of STalign, a software that enables more accurate spatial transcriptomics alignment. The project is spearheaded by her PhD students, Kalen Clifton and Manjari Anant, and was recently published in Nature Communications.
Previous alignment methods relied on affine transformation, which has limitations in accurately capturing the complex, non-linear nature of tissue deformation.
"Our method builds on the Large Deformation Diffeomorphic Metric Mapping algorithm, an alignment technique used in computational anatomy to enable non-linear transformation and smoother alignment with regularization constraints,” she said. “We also added other optimization techniques, such as rasterization of the spatial cell position data to allow for more time efficient alignment."
Fan shared a recent project that compared SRT data of normal and ischemic kidneys. When evaluating at the whole tissue level, the team initially found that gene expression levels appeared similar across samples. However, further comparative spatial analysis using SRT and STalign highlighted spatial pattern alterations of T lymphocyte cells. The finding underscores the importance of spatial analysis in understanding the complexities of gene expression in disease pathology.
"From these, we can see if there are any particular genes or pathways that may allow us to better understand ischemia in kidneys or potentially identify new biomarkers that allow us to better stage the severity of kidney injury," Fan shared.
Reflecting on the challenges encountered throughout her research journey, Fan acknowledged the occasional gaps between statistical models and their real-world applicability.
"One problem that needs to be addressed by the field is how to develop algorithms that are robust against noises and have a low false positive rate," she said. "Even if our statistical models identify cell types that are near each other or are expressing complementary receptor ligand genes, we are still making a jump in concluding that they are interacting... Until we validate with experiment, we don’t know for sure."
Concluding the interview, Fan shared positive prospects toward the future applications of SRT in healthcare.
"Right now, a single SRT experiment still costs thousands of dollars. By analyzing this data, we aim to identify the most useful genes for diagnostic purposes so that commercialization will be possible at a more affordable price," Fan said.