Researchers, students and faculty attended the most recent installment of the Hopkins Department of Chemistry Colloquium Seminar Series last week. Working at the interface of biology and chemistry, Shu-ou Shan, Altair professor of chemistry at the California Institute of Technology, gave a seminar detailing the methods cells use to create “order from chaos.”
Titled “Nascent Protein Selection and Triage at the Ribosome,” Shan’s talk focused on the molecular mechanisms cells employ to selectively create different proteins out of the biochemical goop found in cells.
According to Shan, by understanding the ways in which cells can create proteins, scientists gain incredible insights into understanding how cells “respond to environmental cues and is essential toward understanding processes such as proliferation, differentiation and death.”
As Shan explained, one question is how cells “know” to perform these incredibly specific reactions.
“You look into the textbook, and the cell is perfect. Every cellular pathway knows, what molecular actions should happen, when and where they happen and which order or sequence they happen in,” she said.
Shan explained that problems arise when you leave the idealized world of the textbook to look at simulations of cells as they operate in the real world, with all of their stochasticity and variance.
“How do we generate a high degree of automation and fidelity out of this chemical chaos?” she asked.
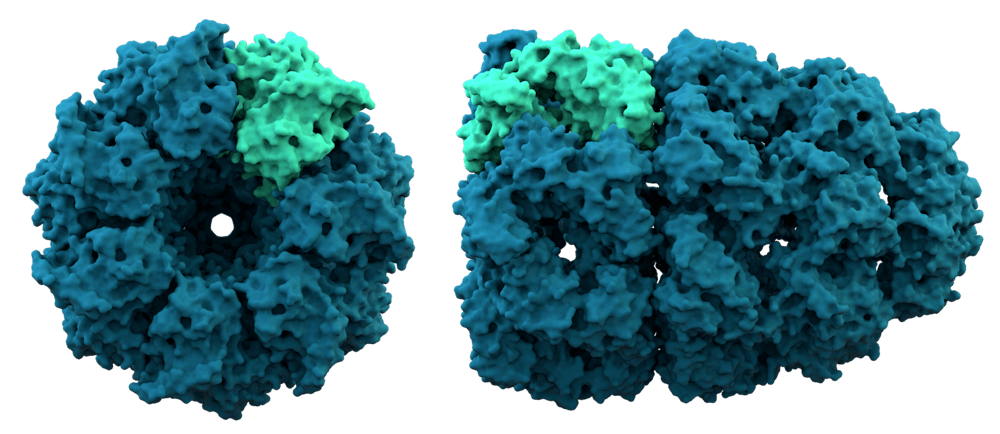
To answer this question, Chan and her team turned to the ribosome, the site of protein synthesis in cells. Ribosomes perform a variety of different functions and can be seen as a kind of hub from which molecular synthesis takes place, having roles in protein folding, localization and quality control.
What Shan’s team keyed on in, however, was the role of ribosome-associated protein biogenesis factors (RPBs), which have been previously shown to play key roles in the early stages of protein synthesis.
What they found centered in large part on two distinct modalities employed in the coordination by RPBs in nascent protein synthesis. To put it shortly, two factors within RPBs have been seen to have a relatively large impact on the early stage of protein synthesis. The first is the signal recognition particle (SRP).
SRPs are ribonucleoproteins that work to shuttle molecular materials to the endoplasmic reticulum (ER), another part of the cell wherein ribosomes are located and proteins can be made.
SRPs, as Shan explained, have been implicated in the transportation of over 30% of the proteome to the ER.
However, once more the question can be raised in regards to fidelity: How does the SRP have such high specificity in helping to carry these intricate materials to the ribosome to be synthesized? As Shan explained, kinetic studies have shown that SRP-dependent protein targeting is highly specific but that purified SRPs have less propensity toward rejection of incorrect biomaterials, begging yet another question.
“Are we losing something in this system? Is there another factor that contributes toward the recognition of proteotic material toward molecular recognition?” she asked.
Enter nascent-polypeptide-associated complex (NAC). NAC is a chaperone protein that has been shown to have considerable import in protein targeting mechanisms. Shan explained that it is integral to preventing the mistargeting of mitochondrial proteins to the ER. And it is this interplay between the NAC and SRP that contributes toward the ability of cells to enact these broadly complex and intricate cellular pathways with such a high degree of specificity.
Shan would also speak on the molecular machinery responsible for shuttling proteins to and from the cytoplasmic membranes of cells, namely the importance of the Sec translocon in interacting co-translationally with the ribosome. This is essentially speaking to the importance of Sec in transporting nascent proteins to and from the ribosome. As Shan explained, SecA engages with ~50% of nascent secretory proteins, illustrating its importance in conveying the seemingly random assortment of molecules within a cell into a coherent system.
Shan ended explaining how all the systems mentioned above can be collated into a broader model that attempts to describe protein transport within bacterial cells.
“What I like the most about this model is how it employs a diverse system of targeting pathways and protein chaperones in the cell, which together take into account a variety of key parameters such as robustness, specificity and efficiency,” she said.