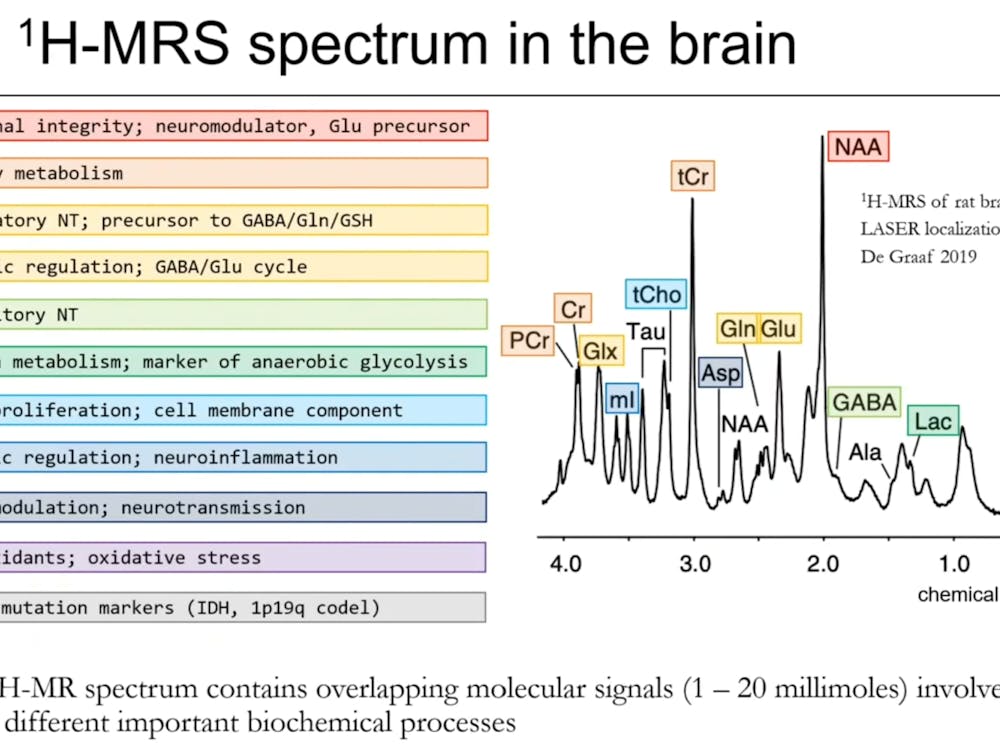
The Hopkins Division of Health Sciences Informatics held its second grand round of the academic year on Oct. 8 about the role of clinical metadata in combating COVID-19 .
Dr. Stuart Ray, professor and vice chair of medicine for data integrity and analytics at the School of Medicine, delivered a lecture titled "Clinical Metadata in Pandemic Response: Structural Barriers and Opportunities.” Throughout his talk Ray discussed his work to derive insight from the large troves of clinical data during COVID-19.
Ray first addressed some of his involvement in studying the genomic diversity of COVID-19 in the early months of the pandemic. His research addressed the following questions in the context of severe acute respiratory syndrome coronavirus two (SARS-CoV-2): What the genetic diversity of the virus is in Baltimore and D.C.; how to measure strains in individuals; and how the strains move among the population.
“We used genomic sequencing to understand the initial spread of SARS-CoV-2 in Baltimore and throughout the U.S.” Ray explained. “By correlating genetic information to disease phenotype, we also aimed to gain insight into any correlation between viral genotype and case severity or transmissibility.”
Ray, along with the rest of the team at Hopkins, analyzed 620 samples collected from the Hopkins Health System between March 11 to 31. The samples amounted to 37.3% of the total cases in Maryland during this period. From these partial genomes, the team produced 114 complete viral genomes. The team discovered that these genomes belonged to all five major clades of SARS-CoV-2. The number of introductions early in the epidemic negatively affected efforts to control local spread of the virus.
In order to sequence the virus, the team used the ARTIC network, an organization that provides a set of materials to assist groups in sequencing the virus. Materials include a set of primers, laboratory protocols, bioinformatics tutorials and datasets.
“The variance in genomes of SARS-CoV-2 suggested multiple introductions of the virus in Baltimore,” Ray said. “We also found that clinically severe cases had genomes belonging to all of these clades.”
Ray and his team are now primarily focused on studying mutations of SARS-CoV-2, which have the potential to make the virus more infectious and dangerous.
There is one mutation in particular involving the spike protein that is alarming to Ray. This spike protein appears to be responsible for stabilizing the fusion polypeptide of the virus once an individual is infected. Increased stability of this spike protein is correlated to a higher infectivity of SARS-CoV-2 and occurs in the mutated G614 spike protein. The original form of the spike protein was the D614 variant.
“The first cases of COVID-19 worldwide showed the D614 variant almost exclusively. Now the G614 variant has almost completely replaced the D614 variant,” Ray said.
The G614 is associated with potentially higher viral loads and infectivity of COVID-19 patients but not with disease severity. Once more of the global population has developed neutralizing antibodies for the G614 protein, Ray suggests we may see a shift back to the D614 variant, which could lower infection rates.
The biggest questions that remain to be answered include how SARS-CoV-2 may be evolving to become more pathogenic and how it may be able to evade antivirals and potential vaccines in the future. Already it has been reported that individuals within the U.S. have contracted COVID-19 multiple times, suggesting a high rate of mutation which the body is not able to provide immunity for.
This question has great relevance to undergoing efforts to produce a vaccine for COVID-19 that is effective over the long term. At the current stage it is unknown for how long a COVID-19 vaccine may provide immunity.
Sophomore Arshdeep Singh felt the event provided hope for the future.
“Hopkins has collected a vast array of genomic samples, which has revealed much of how the virus infects people and causes sickness,” Singh said. “Now their monitoring of mutations of SARS-CoV-2 will allow the world to better prepare for the future of this virus and hopefully develop vaccines that will remain effective for long periods.”