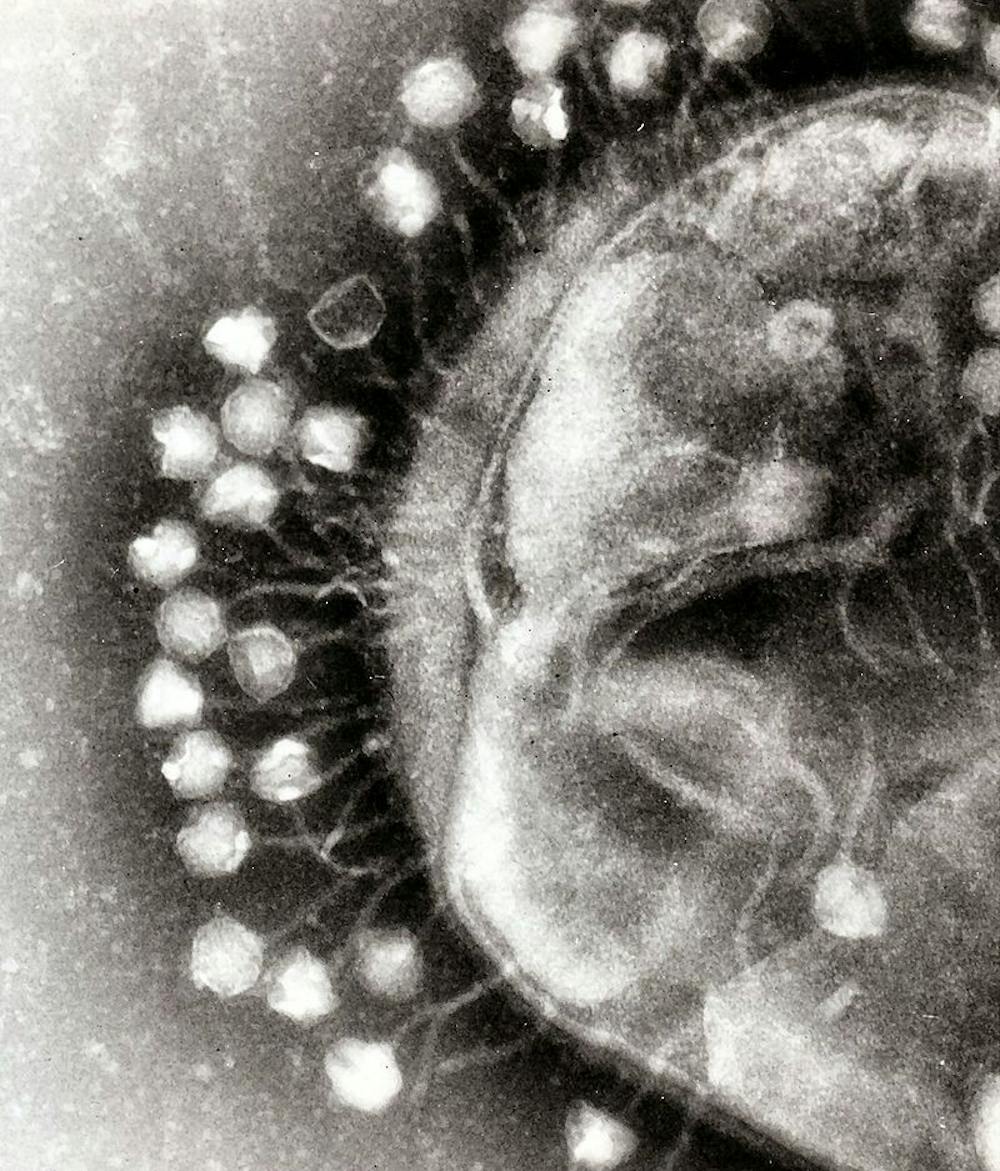
It might be difficult to imagine essentially inanimate objects having a sense of brain and self, but this is virtually the case for viruses. Viruses occupy a gray area. They are generally believed to hover between living and nonliving. Their main purpose for existence is to invade hosts as a sort of intracellular parasite.
Recently researchers at the Texas A&M AgriLife Research and Extension Center found that viruses, just like humans, have a system of competition as well as a criterion of selection.
Viruses, specifically the lambda phages that have been put under scientific investigation, possess a kind of decision-making mechanism. A phage is essentially a type of virus that chooses to replicate inside a bacterium.
Although scientists have been aware of the existence of phages for nearly 100 years, it wasn’t until recently that they began to discover that phages have the potential to attack virulent bacterial species that have become resistant to antibiotics.
This means that viruses could provide a new solution to antibiotic-resistant bacterial diseases. The diversity and vast amount of phages will however contribute to the complexity behind scientists’ efforts to find a cure for bacterial infections and illnesses.
In the journal Nature Communications, Lanying Zeng and her team at Texas A&M AgriLife Research published a paper detailing how the lambda phage chooses to attack its host, the E. coli bacterium. The lambda phage’s tendency to destroy E. coli bacteria causes the two species of the phage to be ideal research targets for Zeng’s team.
Jimmy Trinh, a graduate student working in Zeng’s lab, also studies this interaction closely. Using a four-color fluorescence report system that he designed on his own, Trinh is able to accurately track the lambda phage down to each individual virus.
This technique has been proven useful, especially given the lambda phage’s microscopic size that oftentimes makes it difficult to study. The four-color fluorescence reporter system gave the team insight about phage evolution.
“From the evolutionary point of view, the phages want to optimize their own fitness or survival,” Zeng said in a press release.
In addition, Trinh’s method was joined by Gábor Balázsi’s computational techniques. Balázsi currently works as a biomedical engineer and collaborator at Stony Brook University, a private research institution located in Stony Brook, New York. Trinh’s and Balázsi’s combined efforts allowed them to achieve the primary goal of studying the stimulus or motive that causes phages to destroy their host cells, as well as the phages’ interactions between one another.
“Each phage DNA within the cell is capable of making a decision,” Zeng said. “We want to know how they make a decision, whether one is more dominant than the other, whether they have any interactions and compete to see who will win, or whether they compromise.”
The discoveries made using the four-color fluorescence report system were not what the scientists had initially expected. They found out that lambda phages, despite being far less complicated organisms than humans, also know how to compete or cooperate depending on the situation.
The complexity behind phages’ decision-making can be further explained through their reproductive strategies. The lytic and lysogenic cycles are two ways in which viruses reproduce.
In the lytic cycle, the virus replicates within the host cell but eventually destroys it completely by bursting it open. In the lysogenic cycle, however, the virus is capable of coexisting with its host by inserting its DNA and integrating it into part of the bacterial genome. According to the scientists, the lytic cycle is typically associated with competition while the lysogenic cycle is associated with cooperation.
The discoveries pioneered by Zeng’s team rebuked the stereotypical view of viruses as unintelligent infectious agents.
Currently, Zeng’s team is still seeking to understand more about the viral decision-making process on a cellular level so that their discoveries can potentially be integrated into the cure for antibiotic-resistant bacterial strains in the future.